Drug Discovery and Formulation
1. Introduction
We would like to introduce new features of J-OCTA for the life science and drug discovery fields.
J-OCTA is "multi-scale simulation software" that can calculate phenomena from the atomic and molecular scale (nanometer) to the mesoscale (micrometer) region on the computer. It is based on the free software OCTA, which was originally developed as a Japanese national project to realize multiscale simulations for soft matter (soft materials).
When we think of soft matter, we tend to think of "materials" such as resins and rubbers, but biomolecules and biomembranes are also included in its scope. Recently, we have received an increasing number of inquiries from J-OCTA users related to modeling for life sciences, i.e., drug discovery and drug formulation. In addition, the need for simulation of biomaterials has also been increasing in recent years. In response to these needs, we are expanding J-OCTA's functionality, some of which are introduced below.
2.Simulation of drug formulation
Dissipative Particle Dynamics (DPD), a mesoscale simulation method, can be used to simulate the formation of lipid bilayers and mixed lipid vesicles (liposomes). Vesicles can be used as Drug Delivery Systems (DDS) and have been applied to coronavirus vaccines as Lipid Nanoparticles (LNPs).
Figure 1 shows the shape of LNPs calculated by DPD. Amphiphilic molecules (a single molecule with hydrophobic and hydrophilic ends, respectively) spontaneously form aggregates in solution, with red indicating hydrophilic groups and blue hydrophobic groups. The cross-sectional view shows that hydrophilic groups are also gathered at the center of the sphere. In this area, there are different components in yellow and green. It is to be filled with drugs and other substances.
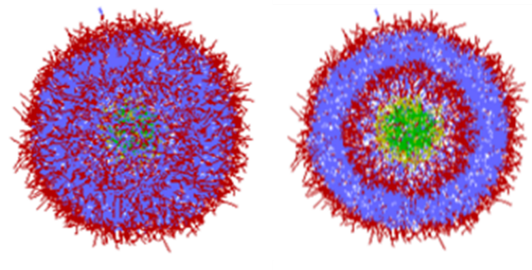 Figure 1. Lipid Nanoparticle (LNP) calculated with DPD (Right : cross section)
Figure 1. Lipid Nanoparticle (LNP) calculated with DPD (Right : cross section)
For details, please refer to the following references [1-3]. The tool used in these references is FCEWS developed by Mochizuki's laboratory at Rikkyo University. The details of the tool are summarized in reference [2], but it is capable of obtaining interaction parameters (called χ (chi) parameters) between coarse-grained units used in DPD using quantum chemical calculations.
In addition to the "nanoparticle formulations" described above, there are also examples of applying a similar DPD approach to the miscibility of "solid dispersion formulations," etc. Please contact us for more details.
3.Simulation of Drug Discovery
J-OCTA provides an interface to GENESIS[4][5][6], a molecular dynamics (MD) engine. GENESIS is molecular dynamics software developed mainly by RIKEN and distributed as free software (LGPLv3). GENESIS Windows version is bundled with J-OCTA. GENESIS supports large-scale parallel computing and GPUs, enabling fast computation of large models. GENESIS also has excellent features such as efficient search for binding structures of proteins and ligands (drugs) using the replica exchange method and free energy calculations to evaluate binding affinity.
J-OCTA offers a new GENESIS Modeler for biomolecular modeling and result analysis. After obtaining the molecular structure of a protein using a database such as PDB or, more recently, AI technology such as AlphaFold2, the force field parameters are set to be suitable for biomolecule calculations. We then apply ionization and other processing to support MD calculations. After the calculation, the user can easily perform various result analyses on the GUI as well as drawings (Figure 2).
In addition, recently there is a demand for the application of simulations not only to small molecules, but also to mid- to high-molecular weight pharmaceuticals. In this case, calculations of large systems are required. J-OCTA has long supported multiscale modeling and results analysis of large systems, and GENESIS' strengths are also demonstrated.
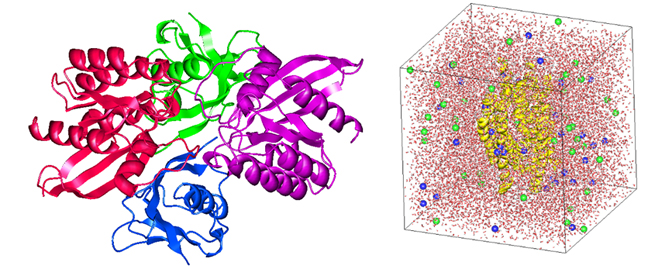 Figure 2. Protein modeling by J-OCTA’s GENESIS Modeler
Figure 2. Protein modeling by J-OCTA’s GENESIS Modeler
This field, called In Silico Drug Discovery, requires a variety of methods and know-how, including AI, data science, docking calculations, and molecular simulations such as MD. Since it is difficult for J-OCTA to handle everything immediately, we will provide functions from our areas of expertise such as multi-scale simulation.
4.Conclusion
We have introduced new functions of J-OCTA for the life science and drug discovery fields. These functions can also be used for simulation of biomaterials such as force field parameters and molecular modeling of glycans.
If you have a topic of interest, please feel free to contact us.
- Reference
- [1] Chemical Physics Letters, 684, 427, (2017),
http://www.sciencedirect.com/science/article/pii/S0009261417307029 - [2] J. Phys. Chem. B, 122, 338, (2018),
http://pubs.acs.org/doi/abs/10.1021/acs.jpcb.7b08461 - [3] Advanced Functional Materials, 30, 1910575, (2020),
https://onlinelibrary.wiley.com/doi/full/10.1002/adfm.201910575 - [4] https://www.r-ccs.riken.jp/labs/cbrt/
- [5] J. Compute. Chem. 38, 2193-2206 (2017),
http://dx.doi.org/10.1002/jcc.24874 - [6] WIREs Comput. Mol. Sci., 5, 310-323 (2015),
http://dx.doi.org/10.1002/wcms.1220


