Materials & Process Informatics
- Machine Learning for Materials Design in J-OCTA -
1. Introduction
The latest trends in machine learning related technologies used by J-OCTA are introduced. New technologies, such as the ability to obtain molecular structures from physical properties, and the ability to predict molecular motions over long time from short-time molecular dynamics results, are included. Please also refer to our previous case study [1] for the materials informatics.
An overview is summarized in Figure 1.
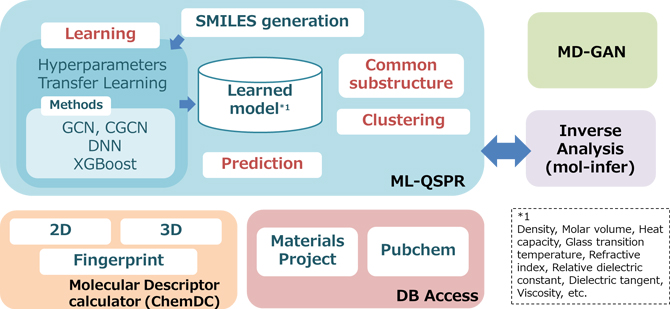 Figure 1. Overview of J-OCTA's machine learning capabilities
Figure 1. Overview of J-OCTA's machine learning capabilities
2. Prediction of physical properties from molecular and crystal structures
In a previous article [1], we introduced the QPSR function using GCN (Graph Convolutional Network). This function learns and predicts the relationship between molecular structures represented as graphs and physical properties. Recently, we have added more functions, which are introduced below.
First, we have increased the number of physical property DBs and learned libraries included with the software. The physical properties shown in the lower right corner of Figure 1 are now supported. To support inorganic crystals, we have also added support for a method called CGCN [2]; by using GCN and CGCN, it is possible to predict physical properties based on molecular and inorganic crystal structures.
Next, we have added support for DNN (fully-connected deep neural network) and XGBoost (a type of decision tree model) as methods other than GCN, which use only molecular structures as explanatory variables (inputs), while DNN and XGBoost can be used for more general applications. For example, ChemDC (a descriptor calculation function based on RDKit[3]), also included in J-OCTA, can be used to obtain molecular descriptors, and by combining this data with other conditions as explanatory variables (inputs), it is possible to predict properties that take into account, for example, process conditions other than molecular structure (Process Informatics). More general use is also possible without using molecular descriptors as explanatory variables (inputs).
Machine learning requires a large amount of physical property data. As a way to obtain physical property values, we introduced the Modeling API function in a previous article [1]. This makes it possible to acquire a large amount of physical property data through high-throughput simulations. On the other hand, it also allows data acquisition from publicly accessible DBs. As shown in Figure 1, data acquisition from Materials Project and PubChem is available, enabling highly accurate prediction of physical properties based on more physical property value data.
3. Prediction of molecular structure from physical properties
The functions introduced in Section 2 were technologies for predicting physical properties from molecular and crystal structures. In this chapter, we introduce the inverse analysis, i.e., a method to predict molecular structure from physical properties.
J-OCTA has interface with mol-infer, which is being developed in Nagamochi Laboratory of Kyoto University [4]. The procedure is the same as in 2 above, but first, physical properties are predicted from molecular structures using an Artificial Neural Network (ANN). Next, the inverse operation of the ANN is solved by Mixed Integer Linear Programming (MILP), which enables us to perform fast and accurate operations in the reverse direction, which is not possible with ANN alone. In this case, a graph structure that seeds the molecular structure and tree structures which correspond to the functional groups are prepared, and these are used to predict the molecular structure.
Figure 2 shows the partition coefficient data used in the test calculations. First, the relationship between molecular structure and physical properties is trained using 1297 data. Next, we perform the inverse operation using MILP. Targeted partition coefficient is 10.0.
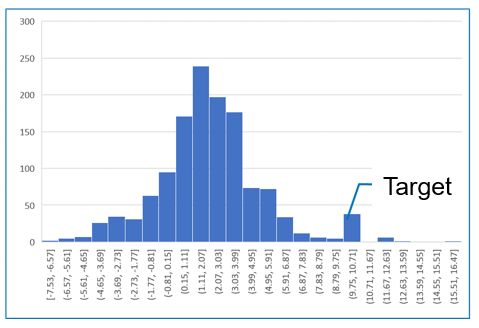 Figure 2. mol-infer's training data and target property ( partition coefficient = 10.0)
Figure 2. mol-infer's training data and target property ( partition coefficient = 10.0)
The resulting structure is shown in Figure 3. Here, structural isomers were also obtained. For the obtained molecular structure, the forward physical properties were again estimated, and the value of the partition coefficient was found to be 9.8. The obtained molecular structure was shown to almost satisfy the target physical property.
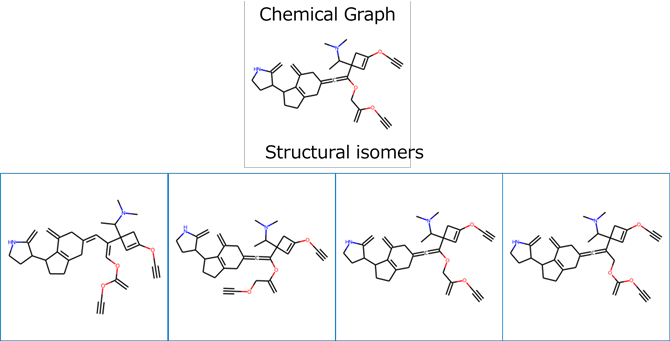 Figure 3. Molecular structure obtained by mol-infer
Figure 3. Molecular structure obtained by mol-infer
4. Machine learning to assist Simulation
Up to the above, machine learning has been used to connect the relationship between physical properties/processes and molecular/crystal structures, but machine learning can be applied to other areas as well.
J-OCTA includes MD-GAN, which predicts the dynamics of a molecule over a long time period based on the results of short-time Molecular Dynamics (MD) calculations[5]. This is an example of the use of machine learning to assist in simulation. Other examples of the use of machine learning include force field calculations (molecular interactions), but are not yet included in J-OCTA. If you have any requests, please contact us.
MD-GAN is a technique developed in the Yasuoka Laboratory at Keio University. For details, please refer to the case study page [5]. MD-GAN can be used to predict long time regions of Mean Square Displacement (MSD), as shown in Figure 4. In this figure, the dotted data in the green region was used to predict the red data for the entire region. You can see that the white long-time region agrees well with the reference data. The effect is difficult to see in simple systems, but when applied to complex phenomena such as polymer melts or the diffusion of Li ions in all-solid-state batteries.
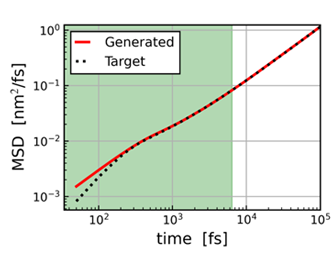 Figure 4. MSD obtained with MD-GAN
Figure 4. MSD obtained with MD-GAN
5. Conclusion
New technologies using machine learning are constantly evolving, and J-OCTA will focus on implementing technologies that we consider useful for users, so please feel free to contact us if you are interested or have any requests.
6. References
- [1] https://www.j-octa.com/cases/caseA49/
- [2] Phys.Rev.Lett.,120,145301,(2018)
- [3] RDKit:https://www.rdkit.org/
- [4] Int.J.Mol.Sci.,22,2847,(2021)
- [5] https://www.j-octa.com/cases/caseA52/


