- Quantum Chemistry / DFT
- DPD / Mean Field
- Multiscale Analysis
- Interface / Phase Separation / Particle Dispersion
- Life Science
[Analysis Example] Formation of lipid bilayers and vesicles using FMO-DPD
Prediction of χ parameters with FCEWS, and Mesoscale analysis of phospholipids
Dissipative Particle Dynamics (DPD), a mesoscale simulation method, can be used to simulate the formation of lipid bilayers and vesicles (liposomes) of mixed lipids. Vesicles can be used as lipid nanoparticles (LNPs) in drug delivery systems (DDS).
In the following example, the structure of a typical phospholipid, POPC, forming in water was simulated using DPD [1].
First, the lipid molecule is divided into segments (Figure 1), and the χ parameters (interaction parameters) between the segments are estimated by FCEWS, which can predict χ parameters with high accuracy by using the Fragment Molecular Orbital method (FMO). Four water molecules are considered as one DPD particle.
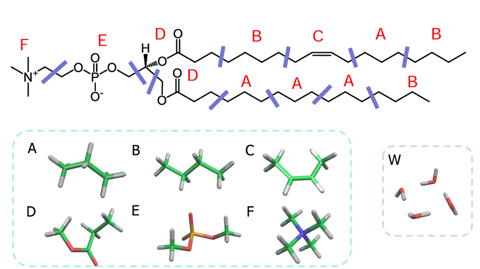 Figure 1. Segmentation of POPC molecule and water
Figure 1. Segmentation of POPC molecule and water
Mesoscale simulations were performed using DPD. By varying the lipid fraction we can see the formation of lipid bilayers and vesicles (Figure 2). Analysis of the lipid bilayer results shows that the area per lipid , which represents the area occupied by a single lipid molecule, is 69.4 Å2, while the experimental values are in good agreement, ranging from 62.7 to 68.3 Å2. The thickness of the lipid bilayer is 2.58 nm, while the experimental value is 2.8 nm, also in good agreement.
FCEWS is developed by Mochizuki lab. of Rikkyo Univ., and included in J-OCTA. For more details, please refer to references [1][2].
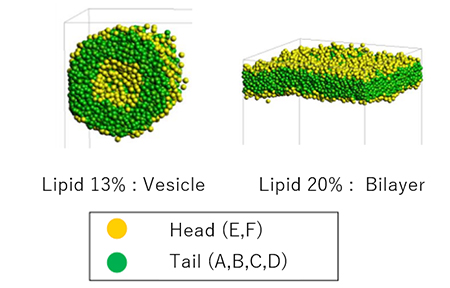 Figure 2. Structures in water calculated by DPD.
Figure 2. Structures in water calculated by DPD.
- Reference
- [1] Chem.Phys.Lett., 684, pp427-432, (2017)
- [2] J.Phys.Chem.B, 122, pp338-347, (2018)


