- Data Science
- Small molecule penetration / diffusion / adsorption
- Material Science
[Analysis Example] Analysis of Li ion diffusion in solid-state battery by MD-GAN
Long time series data generation using a deep generative model based on AIMD trajectories
Molecular Dynamics (MD) are effective in providing insights at the atomic level, However, the computational costs are notably high, particularly ab initio MD (AIMD). Even as parallelization techniques have expanded the spatial scale of MD, the extension of the temporal scale remains a persisting challenge. MD-GAN[1,2,3] is a machine learning model using Generative Adversarial Network (GAN) capable of generating long-time data from multiple short-time data segments. In other words, it enables parallelization of temporal calculation.
In this study, MD-GAN is trained on Li ion diffusion in solids calculated by AIMD to generate long-term time series MD data.
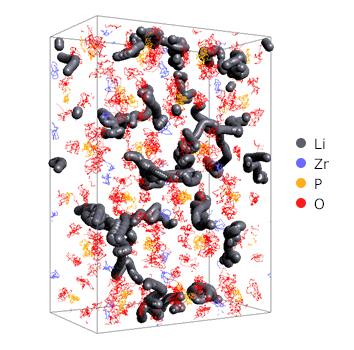 Figure 1. visualization of ion paths (1ps)
Figure 1. visualization of ion paths (1ps)
Figure 2 shows an overview of MD-GAN.
In input data preparation, particles of interest (Li ions) are extracted from the MD data.
MD-GAN learns the rules for 128 records in the input data. The time interval between records is determined by step skip, and the length of a time series to be trained at a time is 128×step skip; the smaller the step skip, the more samples that can be taken from the same input data.(Figure 3)
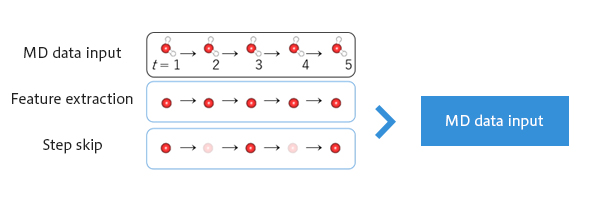 Figure 2. Overview of MD-GAN
Figure 2. Overview of MD-GAN
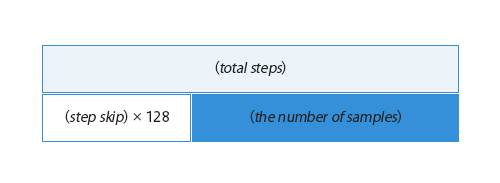 Figure 3. Relationship between step skip and the number of samples
Figure 3. Relationship between step skip and the number of samples
In this case, we performed 3 MD simulations for LiZr2(PO4)3, which is a material used in solid-state electrolytes. Step skip was set to 2, and the total steps was set 25000 in MD-GAN.
Table shows the details of the calculation conditions.
The number of Li ions in the input data was 12 × 3, whereas the number of Li ions in the generated data was 8192.
We calculated and compared the mean square displacement (MSD) and coordinate displacement for MD and MD-GAN, respectively.
Figure 4 shows the displacement of the z-coordinate of each particle in both MD and MD-GAN. MD-GAN successfully replicated the random walk observed in MD.
Figure 5 illustrates MSD of MD data and MD-GAN. The gray area represents the duration of the training data. The time-series data are generated with high accuracy, matching nonlinear regions with small time scales to regions larger than the learned time-series data.
Table Calculation conditions for MD and MD-GAN
MD simulation conditions
| Solver | AIMD with SIESTA[4] |
|---|---|
| Total steps | 10000 |
| Delta t | 4fs |
| Temperature | 1773K |
| Number of trajectories | 3 |
MD-GAN conditions
| SStep skip | 2 |
|---|---|
| Feature extraction | Li ion |
| Extracted particles | 12 |
| Input particles | 12×3 |
| Generated particles | 8192 |
| Machine | Computation time | |
|---|---|---|
| MD | CPU (Intel(R) Xeon(R) Gold 6226R CPU @ 2.90GHz) | 30 days |
| MD-GAN | GPU (NVIDIA Quadro T1000) | 28 hours |
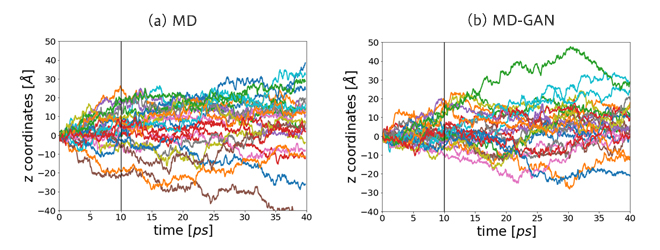 Figure 4. z-coordinate displacement of Li ions in (a)the MD data and (b)the time-series data generated by MD-GAN
Figure 4. z-coordinate displacement of Li ions in (a)the MD data and (b)the time-series data generated by MD-GAN
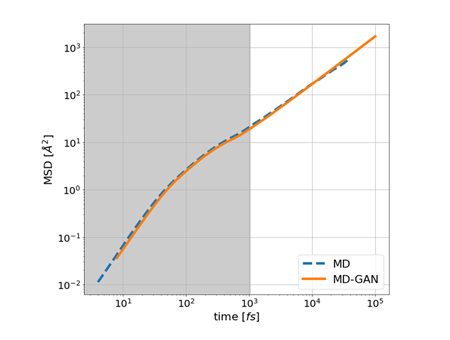 Figure 5. Comparison of MSD in MD data and MD-GAN
Figure 5. Comparison of MSD in MD data and MD-GAN
- References
- [1] Endo, K. et al., Proceedings of the AAAI Conference on Artificial Intelligence, 32, 2192 (2018).
- [2] Kawada, R. et al., Journal of Chemical Information and Modeling, 63, 76-86(2022).
- [3] Kawada, R. et al., Soft Matter, 18, 8446-8455(2022).
- [4] Soler, J. M. et al., J. Phys. Condens. Matter, 14, 2745 (2002).


